New AI Tool Accelerates mRNA-Based Treatments for Viruses, Cancers, Genetic Disorders
UT Austin and Sanofi partner to build tool that predicts translation efficiency of mRNA sequences.

A new artificial intelligence model can improve the process of drug and vaccine discovery by predicting how efficiently specific mRNA sequences will produce proteins, both generally and in various cell types. The new advance, developed through an academic-industrial partnership between The University of Texas at Austin and Sanofi, helps predict how much protein cells will produce, which can minimize the need for trial-and-error experimentation, accelerating the next generation of mRNA therapeutics.
Messenger RNA (mRNA) contains instructions for which proteins to make and how to make them, enabling our bodies to grow and carry out the day-to-day processes of life. Among the most promising areas of health and medicine, the ability to develop new mRNA vaccines and drugs — able to fight viruses, cancers and genetic disorders — involves the frequently challenging process of coaxing cells in a patient’s body to produce enough protein from therapeutic mRNA to effectively combat disease.
The new model, called RiboNN, stands to guide the design of new mRNA-based therapeutics by illuminating what will yield the highest amount of a protein or better target specific parts of the body such as the heart or liver. The team described their model today in one of two related papers in the journal Nature Biotechnology.
“When we started this project over six years ago, there was no obvious application,” said Can Cenik, an associate professor of molecular biosciences at UT Austin, who co-led the work with Vikram Agarwal, head of mRNA platform design data science in Sanofi’s mRNA Center of Excellence. “We were curious whether cells coordinate which mRNAs they produce and how efficiently they are translated into proteins. That is the value of curiosity-driven research. It builds the foundation for advances like RiboNN, which only become possible much later.”
The work was made possible by funding support from the National Institutes of Health, The Welch Foundation and the Lonestar6 supercomputer at UT’s Texas Advanced Computing Center.
In tests spanning more than 140 human and mouse cell types, RiboNN was about twice as accurate at predicting translation efficiency as earlier approaches. This advance may lend researchers the ability to make predictions in cells in ways that could help expedite treatments for cancer and infectious and hereditary diseases.
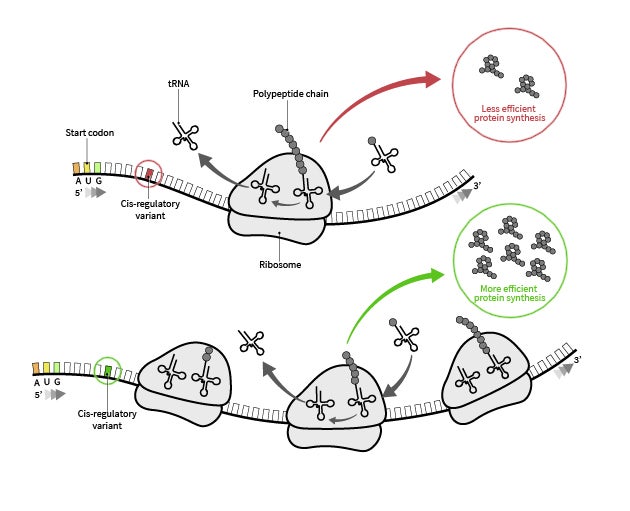
Subtle differences in an mRNA sequence enables a ribosome to produce more or less of a certain protein. A new AI model called RiboNN predicts which sequences will be most efficiently produced and potentially, most effective for protein-based therapeutics. Credit: University of Texas at Austin.
You can think of the way cells in your body make proteins as the way a team of chefs might bake cakes. To cook up a batch of proteins, the chefs in one of your cells (ribosomes) look up the recipe in your own unique protein cookbook (a.k.a. DNA), copy the recipe onto notecards called messenger RNAs (mRNAs), and then combine ingredients (amino acids) according to the recipe to bake up the cakes (proteins).
An mRNA vaccine or therapeutic coaxes these chefs in your cells into making proteins. In the case of a vaccine, they might produce a protein found on the surface of a pathogenic virus or cancer cells, essentially waving a big red flag in front of your immune system to make antibodies against the virus or cancer. In the case of a disorder caused by a genetic mutation, they might produce a protein that your body can’t properly make on its own, reversing the disorder.
Before developing their new predictive model, Cenik and the UT team first curated a set of publicly available data from over 10,000 experiments measuring how efficiently different mRNAs are translated into proteins in different human and mouse cell types. Once they had created this training dataset, AI and machine learning experts from UT and Sanofi came together to develop RiboNN.
One goal of the predictive tool is to one day make therapies that are targeted to a particular cell type, said Cenik, who also is affiliate faculty at UT’s Oden Institute for Computational Engineering and Sciences and a CPRIT scholar, receiving research support from the Cancer Prevention and Research Institute of Texas.
“Maybe you need a next-generation therapy to be made in the liver or the lung or in immune cells,” he said. “This opens up an opportunity to change the mRNA sequence to increase the production of that protein in that cell type.”
In a companion paper also in Nature Biotechnology, the team demonstrated that mRNAs with related biological functions are translated into proteins at similar levels across different cell types. Scientists have long known that the process of transcribing genes with related functions into mRNAs is coordinated, but it hadn’t been previously shown that translating mRNAs into proteins is also coordinated.
UT undergraduate student researchers manually checked available data for accuracy and filled in missing information to create RiboBase, the dataset needed to train the AI model. The teams that collaborated to develop RiboNN included Logan Persyn, a UT graduate student in computer science, and Dinghai Zheng and Jun Wang at Sanofi. UT’s Discovery to Impact office helped facilitate the collaboration between UT and Sanofi by developing a research agreement.