How Breast Cancer Hijacks a Natural Enzyme to Boost Mutations
Kyle Miller and his team discovered a potential new target for drug therapies: structures in our DNA called R-loops.
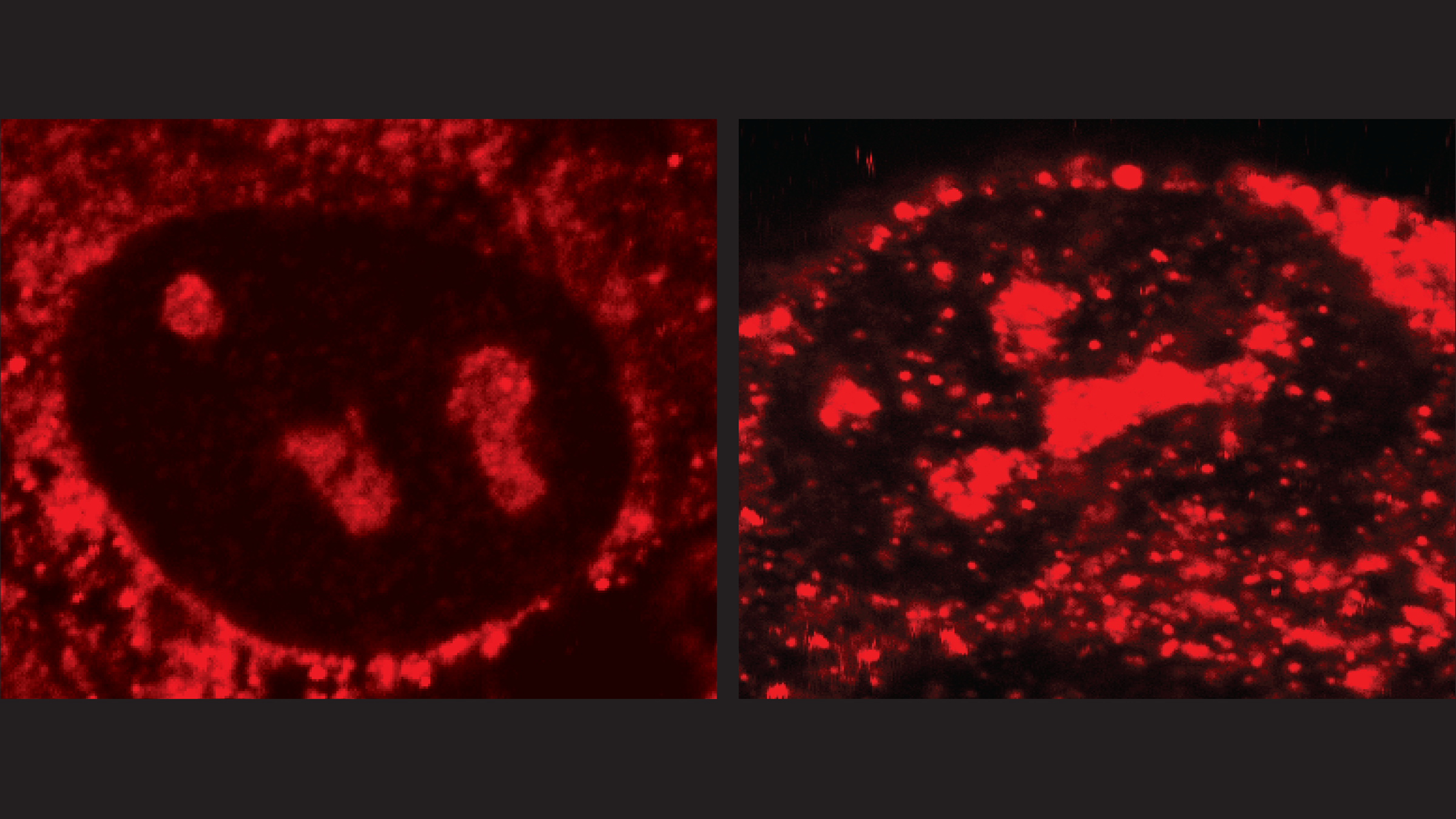
A human cancer cell that overexpresses the enzyme APOBEC3B (left) has fewer R-loops (red) than a cancer cell that has been genetically modified to have no APOBEC3B (right). That’s because the enzyme mutates and then helps resolve R-loops. Credit: Kyle Miller/University of Texas at Austin.
One of the many tricks cancer cells use to outcompete healthy cells is to take an enzyme designed to neutralize viruses and instead use it to randomly tweak the cancer cell’s own DNA. In essence, it enables cancer to roll the evolutionary dice more often, making random mutations that just might, if it’s lucky, give it powerful new abilities. About 20% of breast cancer tumors produce an excess of this enzyme, called APOBEC3B, which is correlated with worse outcomes for patients and more resistance to therapies.
A collaboration of three research teams, including one led by Kyle Miller at The University of Texas at Austin, has discovered that APOBEC3B targets R-loops, sections of a person’s genome that are in the process of being copied to build proteins. This insight could lead to improved methods for fighting some cancers. They published their findings in the journal Nature Genetics.
Most of the time, the DNA in our cells is in the form of a double helix—two strands of DNA wound around each other like two sides of a zipper that’s been twisted. When it’s time to make proteins, a section of the double helix will unzip, forming an R-loop. While one side of the zipper is involved in copying the instructions through RNA for making proteins, the other side is left exposed.
Meanwhile, the enzyme APOBEC3B is circulating around the cell in low levels looking for single-stranded DNA associated with viruses, such as HIV. When the enzyme finds a suspected virus, it makes a small mutation that can neutralize it. Sometimes though, the enzyme mistakes the single-stranded DNA in an R-loop for a virus and attacks it. That happens to a small degree in normal cells, but in cancer it’s turbocharged—there’s a lot more of the enzyme going around, searching for single-stranded DNA—unleashing a cascade of mutations.
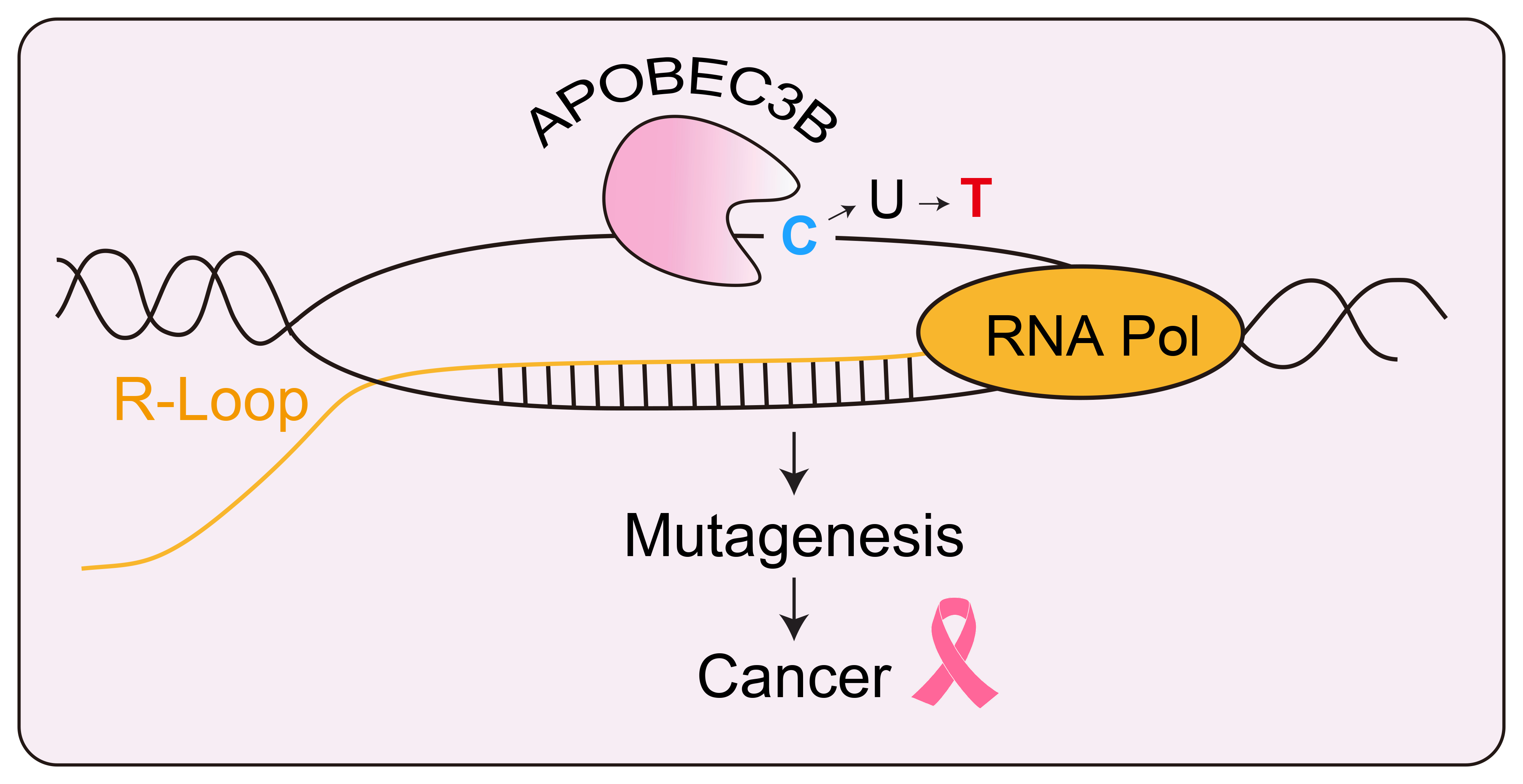
The enzyme APOBEC3B targets the exposed single stranded DNA of an R-loop and changes a letter C to a letter T. In cancer cells, this can happen often and lead to potentially useful mutations. Credit: Kyle Miller/University of Texas at Austin.
“Cancer is an evolutionary process,” said Kyle Miller, professor of molecular biosciences at UT Austin and member of the Livestrong Cancer Institute at Dell Medical School. “Each cell is experimenting and whatever cell is the most efficient is the one that’s going to survive in the tumor and branch out. A therapy might have a great effect on a tumor, but those few cells that are able to mutate and escape that therapy, they’re the ones that become resistant, they’re the ones that metastasize, and those are the ones that kill patients.”
Miller said R-loops could be a potential target for future cancer therapies. A drug that modifies the number of R-loops in a cell’s DNA might act as a counterweight to APOBEC3B.
“This is the same rationale for DNA-damaging agents like radiation and many chemotherapies,” Miller said. “Cancer cells already have lots of DNA damage, so there’s only so much more they can take. Whereas healthy cells have much lower levels to begin with; they can tolerate a little more. So cancer cells are preferentially targeted.”
Essentially, according to Miller, adding R-loops could cause a cancer cell with high levels of APOBEC3B to generate so many mutations that it’s no longer able to carry out its basic functions. In other words, while a certain amount of mutation benefits cancer cells, beyond a certain point, it could become too much. The good news is that there are already known drugs that increase R-loops, some of which are FDA-approved, including topoisomerase inhibitors.
In addition to the UT Austin team—led by Miller, along with two former postdoctoral researchers, Seo Yun Lee and Jae Jin Kim, and one current postdoctoral researcher, Anthony Sanchez—researchers from the University of Oxford (led by the MRC Senior Research Fellow Natalia Gromak and co-first author Agnese Cristini) and from UT Health Science Center at San Antonio (led by Reuben S. Harris and co-first author Jennifer McCann) made the discovery.
The experimental work demonstrating that APOBEC3B targets R-loops was carried out using cancer cells in culture. The researchers compared their observations with medical data from patient tumors (including the expression levels of this enzyme and the levels of associated mutations) and found they were consistent with this interpretation.
Support for this research was provided by the National Cancer Institute, National Institute of Allergy and Infectious Diseases, the Cancer Prevention and Research Institute of Texas (CPRIT) including a Recruitment of Established Investigator Award for Reuben Harris, the Royal Society, the Wellcome Trust, American Cancer Society, U.S. National Institutes of Health, National Science Foundation, Howard Hughes Medical Institute and the Ovarian Cancer Research Alliance. Miller and Reuben Harris are CPRIT Scholars.



